Two wrongs don’t make a right, but sometimes two “broken” genetic mutations can “fix” each other.
That’s what a team of NC State researchers discovered when studying an enzyme involved in making a critical plant growth hormone. Enzymes are proteins made by living organisms to speed up chemical reactions.
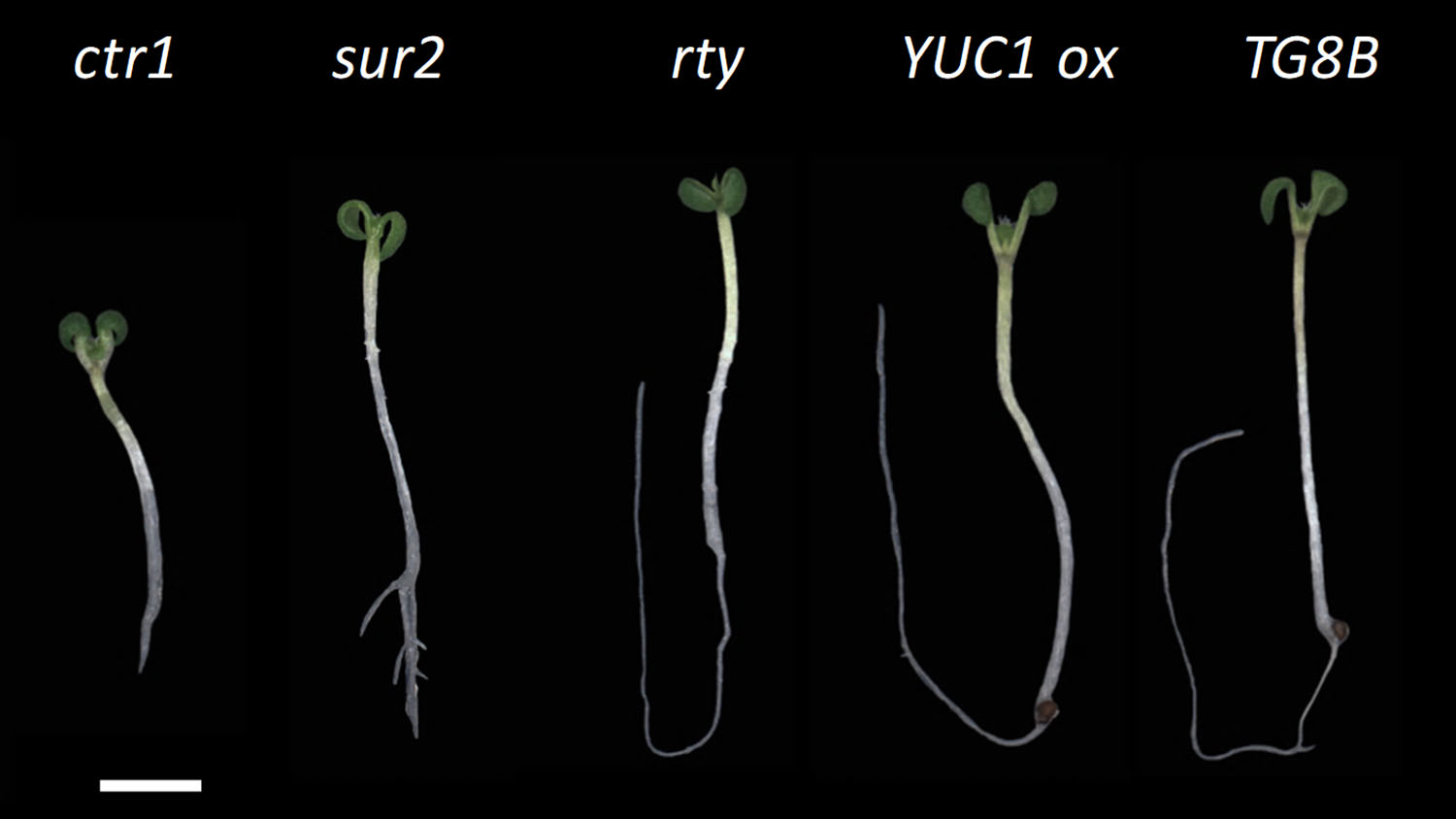
The enzyme is called ROOTY, because the seedlings have characteristic stumpy and branched roots, said Javier Brumos, a researcher in NC State’s Department of Plant and Microbial Biology.
Brumos and his colleagues shared their discovery in a published in the journal Plant Physiology. They found a new mutation in ROOTY that “fixed” a well-studied mutation in the same gene. Then they constructed a computational model to see how the two mutations could fix or complement each other, producing a functional enzyme.
“From the scientific point of view, you’re always looking for novelty, to bring something new to the table,” said Brumos, the first author on the paper. “Our goal was to find new genes that are involved in the production of auxin, a plant hormone. When we identified that the mutation was actually in a well-known gene, we got super depressed. Then we got curious, the mutation is in the same gene: How did it complement the known mutant?”
First Up, Then Down
Brumos and senior author , an associate professor of plant and microbial biology, started with Arabidopsis thaliana, the widely-used (and fast-growing) plant species favored by plant biologists. They generated a massive collection of seeds with random mutations and then looked at 10-day-old seedlings for specific traits. Specifically, seedlings with curly cotyledons, the first two “leaves” of a plant shoot, or short, stumpy roots.
They kept the seedlings with these traits, transferred them to soil, grew them up and bred with themselves. Next, they crossed these “pure bred” mutants with pure bred plants with known mutations in well-studied genes involved in making the plant growth hormone auxin.

“As expected, when we crossed them, most of them looked like the parents, so we knew the new mutation was in the same gene as the known mutation,” Brumos said. “We could discard those mutants because the gene was already known.”
However, there were some normal-looking seedlings from the crosses, which indicated that the new mutant complemented the known mutant.
“Genetics 101 suggested that the mutations were in two different genes,” Brumos said. “We got super excited at that point. We thought we’d found a new gene that was involved in auxin synthesis.”
However, when they mapped and sequenced the new mutation, they were disappointed to learn that the mutation was actually in the well-studied gene, ROOTY.
After they recovered from their disappointment, the team began to get curious as to how two mutations in one enzyme could complement each other, as the findings were rather unexpected.
Many enzymes, including ROOTY, the protein produced by the ROOTY gene, work by pairing up with another copy of the protein to form a functional unit – like two halves of a pair of scissors. The team, with collaborator Douglas Grubb, a group leader at the Leibniz Institute for Plant Biochemistry, had a hypothesis that when a copy of the enzyme with the new mutation paired up with a copy of the enzyme with the well-known mutation, the mis-matched mutant pair was functional. , William Neal Reynolds professor of plant and microbial biology and a long-term collaborator of Stepanova, proposed that the new mutation could disable a different section, or domain, of the enzyme than the well-known mutation.
To test this hypothesis, the researchers teamed up with Ben Bobay, a senior research associate at Duke University, to make a computational 3D model of the ROOTY enzyme – by comparing the normal enzyme to the known structure of “cousin” enzyme, and then overlaying the location of the new mutations and previously discovered mutations.
They found that the new mutation was located where the two copies of the protein met – think the pin that holds a pair of scissors together – while the well-known mutation was in the functional part of the enzyme – think the scissors’ blades. By combining one dull blade with a proper pin and one sharp blade without a pin, you can produce a pair of scissors that could work, abet not very well.
“The beauty of this work is that we did the benchwork to identify the new mutants and then we did computational modeling to test many, many combinations between these mutants,” Brumos said. “We could clearly see that if one of the monomers has the mutation on the interaction surface and the other monomer has a mutation in a different domain, then they were able to come together and form a functional enzyme.”
There are not many examples of this type of complementation in the scientific literature, Stepanova said. Though it likely does occur in nature more frequently than reported.
An example this type of complementation has been reported in the human disorder phenylketonuria, or PKU. PKU is caused by having mutations in the gene that codes for an enzyme that breaks down the amino acid phenylalanine. Unless on a very special diet, phenylalanine can build up to toxic levels in a baby or individual with PKU, causing a host of neurological issues. However, some individuals with different mutations in each copy of the gene have been reported to show milder symptoms, due to this kind of complementation.
In addition to highlighting new areas of research, the paper’s findings should serve as a warning for budding geneticists, Stepanova said.
“This is a cautionary tale for scientists doing this type of complementation testing,” Stepanova said. “When you do this type of test, you’re supposed to use a mutant that cannot make any protein for your control. But sometimes scientists use whatever well-known mutants are available. And if they cross two mutants with mis-sense mutations, there’s a possibility of functional complementation, as we have shown.”
While some of the other mutants identified in the screen have been by Stepanova and her team, work on others is still ongoing and will be published in the future.
In addition to Brumos, Stepanova, Alonso and Bobay, Cierra Clark, an undergraduate student who graduated in May 2019, was involved in the study and was an author on the paper.
The │į╣Ž═°šŠ Science Foundation funded the research.
Our research addresses grand challenges – and overcomes them.
This post was in College of Agriculture and Life Sciences News.
- Categories:
- Tags:








