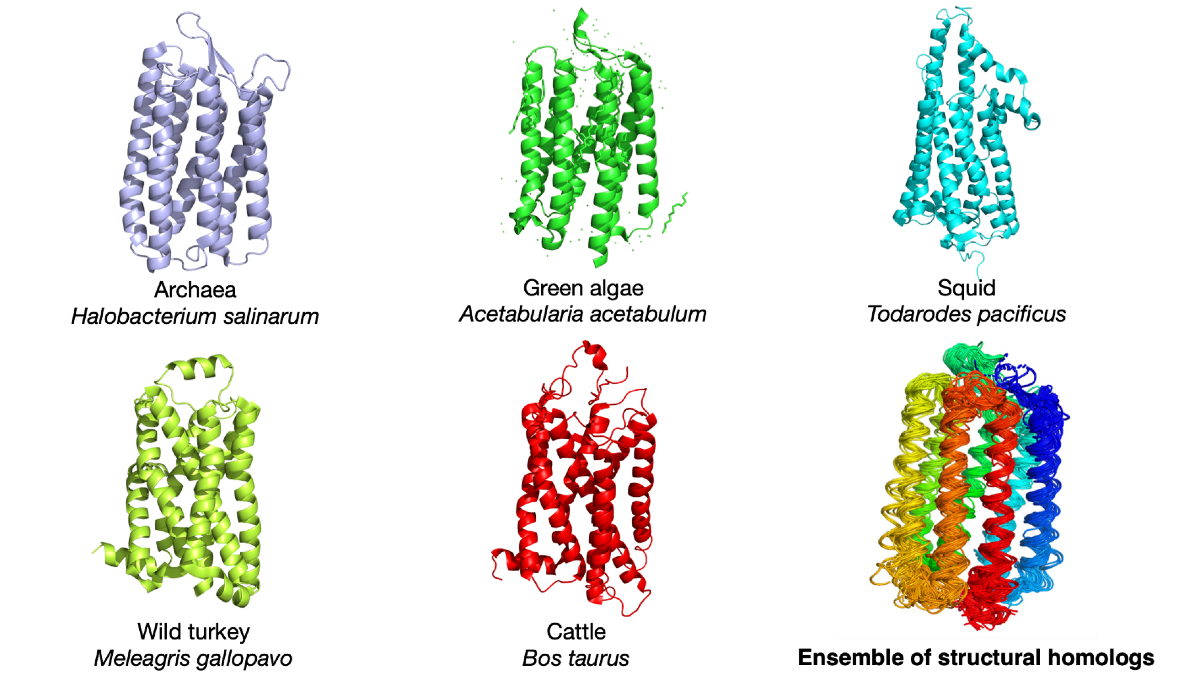
Proteins common in multiple species were compared to understand the relationship between how proteins change shape, both over seconds or minutes and throughout evolution. The image shows the light-detecting protein rhodopsin in five different species as well as an overlay to reveal how the protein structure has changed with evolution. © Qian-Yuan Tang,
A recent statistical study has revealed some of the constraints and directions in the evolution of the structure and function of proteins. Better models of protein structural dynamics may allow researchers to understand more of this fundamental mystery in living organisms.
Proteins perform essential functions such as material transport, immunity and catalysis. The various functions of proteins have evolved gradually over the course of evolution.
Due to thermal noise, or the random motion of atoms, proteins shift their structures while performing their functions. These dynamics usually happen in short time scales, from microseconds (millionth of seconds) to milliseconds (thousandth of seconds). Meanwhile, genetic mutations may also lead to variations in protein structures, leading to the evolution of proteins. Such an evolutionary process happens generation after generation, which correspond to a much longer time scale.
“Interestingly, although being two completely different processes, the dynamics and the evolution of proteins share many similarities. However, it is not easy to verify quantitatively, and the theoretical origin of the relationship has not yet been clarified before,” said Qian-Yuan Tang, Ph.D., co-author of the research published in Physical Review Letters and a former postdoctoral research fellow at the University of Tokyo. Tang is currently a research scientist at the RIKEN Center for Brain Science in Japan.
In the recently published research, Tang and Professor Kunihiko Kaneko, a theoretical biology expert from the University of Tokyo Research Center for Complex Systems Biology, analyzed the structures of hundreds of thousands of proteins in scientific databases. These proteins can be divided into different groups based on their structural similarity. The proteins in the same group are usually the same kind of protein in different animal species. For example, the iron-carrying blood protein hemoglobin in humans, rats and fishes will be in the same group. The structural variations within a group reflect the structural evolution of the proteins. Further analysis shows that the structural variations that happened in evolution and the structural variations that happened in the functional dynamics of proteins share the same pattern.
“What we are finding is a connection between thermal noise-induced deformations and mutation-induced deformations,” said Tang.
“This correspondence between dynamics and evolution is due to the fact that structural changes of proteins due to thermal fluctuations and mutations share the same constraints,” said Kaneko.
These constraints lead to a close relation between the directions in which protein functional movement and structural evolution tend to occur. The emergence of such constraints can be explained by the fact that protein structure must be resistant to thermal noise and genetic mutations, but also must be sensitive enough to function.
A better understanding of this correspondence may provide a unified way to understand the functional behavior of proteins and analyze their evolutionary constraints. These results also give a new perspective to the design of functional living systems and artificial intelligence systems.








