New findings on Sars-CoV-2 protein shed light on virus’ ability to infect cells
In order to infect cells, the virus needs the spike protein on its viral surface. Scientists from the Max Planck Institute for Biophysics, the Paul Ehrlich Institute, the European Molecular Biology Laboratory (EMBL), and the Goethe University Frankfurt, have analysed the spike protein in its natural environment using high-resolution imaging and computer-based methods. In the process, they have gained surprising insights, including an unexpected freedom of movement. The findings could help in the search for vaccines and medicines.
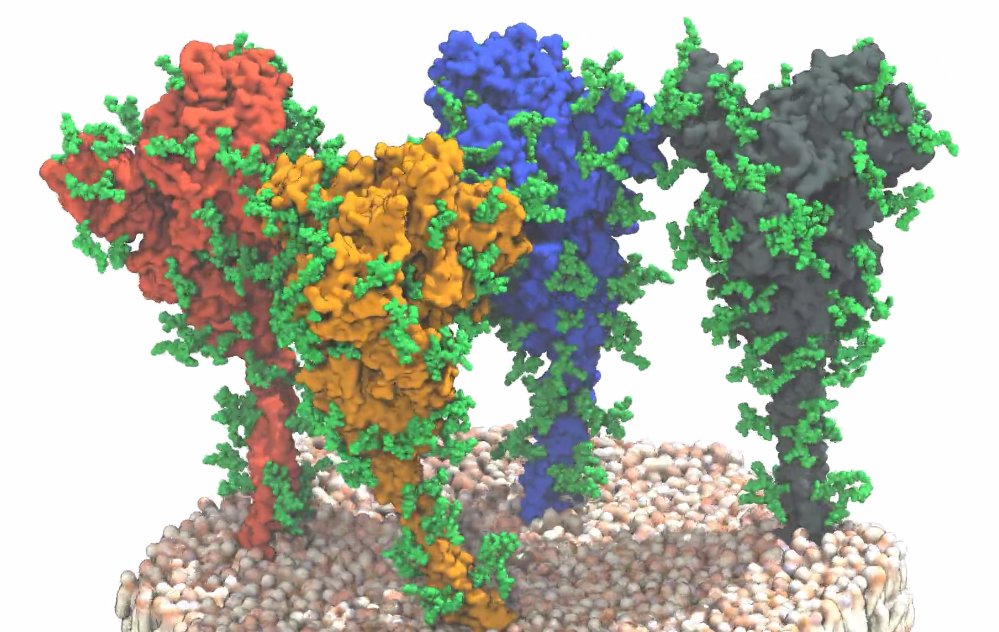
At the start of a Covid-19 infection, the coronavirus Sars-CoV-2 docks onto human cells using the spike-like proteins at its surface. This protein is at the centre of vaccine development because they trigger an immune response in humans. Therefore, a group of German scientists, including members from the European Molecular Biology Laboratory (EMBL) in Heidelberg, the Max-Planck Institute of Biophysics, the Paul Ehrlich Institute, and Goethe University Frankfurt focussed on the surface structure of the virus to gain insights for the development of vaccines and of effective therapeutics to treat infected patients.
The team combined technologies of cryo-electron tomography, subtomogram averaging and molecular dynamics simulations to manage the analyses the of molecular structures of these spike proteins in its their natural environment, on intact virions, and with near-atomic resolution. Using EMBL’s state-of-the-art cryo-electron microscopy imaging facility, 266 cryo tomograms of about 1000 different viruses were generated, each carrying an average of 40 spikes on its surface. Subtomogram averaging and image processing combined with molecular dynamics simulation finally provided the needed important and novel structural information from these spikes.
Flexible stalk
The results were surprising: The data showed that the globular portion of the spike protein, which contains the receptor-binding region and the machinery required for fusion with the target cell, is connected to a flexible stalk. “The upper spherical part of the spike has a structure that is well reproduced by recombinant proteins used for vaccine development,” explains Martin Beck, group leader at EMBL and director at the Max Planck Institute of Biophysics of Biophysics. “However, our findings about the stalk, which fixes the globular part of the spike protein to the virus surface, were new.”
“The stalk was expected to be quite rigid,” adds Gerhard Hummer, from the Max Planck Institute of Biophysics and the Institute of Biophysics at the Goethe University Frankfurt. “But in our computer models and in the actual images, we discovered that the stalks are extremely flexible.” By combining molecular dynamics simulations and cryo tomography, the team identified the three joints – hip, knee and ankle – giving the stalk its flexibility.
“Like a balloon on a string, the spikes appear to move on the surface of the virus and thus are able to search for the receptor for docking to the target cell,” describes Jacomine Krijnse Locker, group leader at the Paul Ehrlich Institute, the results. To prevent infection, these “balloons” are targeted by antibodies. However, the images and models also showed that the entire spike protein, including the stalk, is covered with glycan chains – sugar-like molecules. These chains provide a kind of protective coat that hide the spikes from neutralising antibodies. Another important finding on the way to effective vaccines and medicines.








