Max Planck researchers want to analyse the surface protein of the coronavirus to identify binding sites for vaccines and drugs
The spike glycoproteins give the coronavirus its name. The molecules protrude from the viral envelope like the spikes of a crown. Researchers at the Max Planck Institute of Biophysics in Frankfurt are now analysing the structure of this protein. They hope to identify potential targets for antibodies and inhibitors – an important prerequisite for developing new vaccines and drugs against the SARS CoV-2 virus.
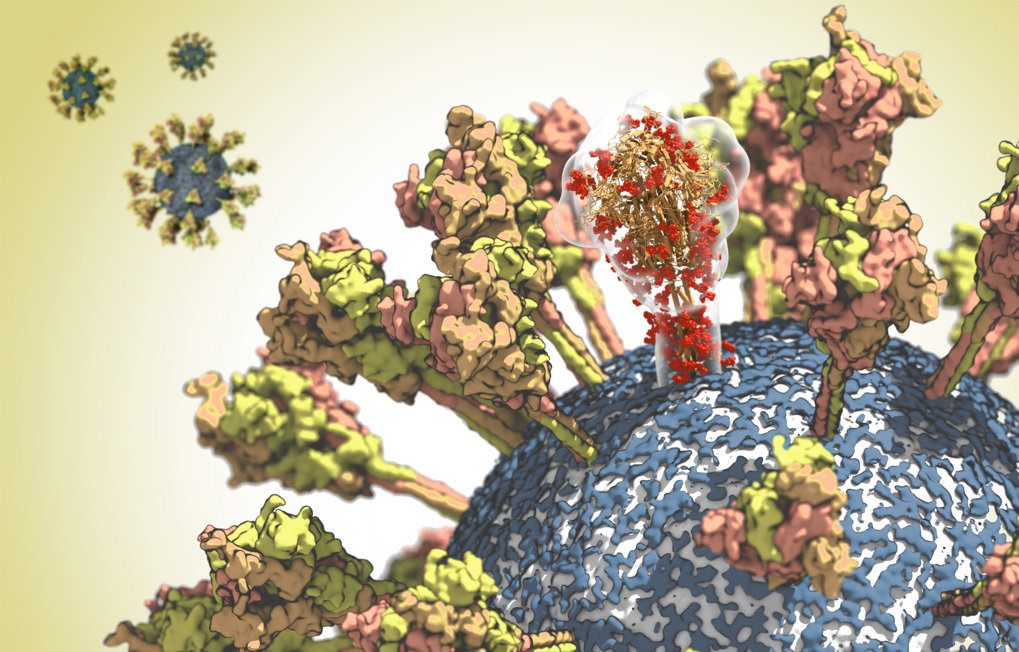
The coronavirus needs the spike protein to infect a cell. The protein binds primarily to a receptor called ACE2 on the surface of human cells. The virus can then fuse with the cell membrane and release its genetic material into the cell.
The spike protein is not only the sharpest weapon of the virus but also its Achilles’ heel; its exposed position makes it the preferred point of attack for the immune system. Antibodies can recognize the virus by its spike protein, bind to it, and thus mark it as a target for immune cells. However, the virus has another trick up its sleeve. A sugar coat hides the conserved parts of its spike proteins from the immune cells.
Protective shield made of sugar
The Max Planck researchers are therefore analysing the protective sugar shield and the membrane envelope of the virus in addition to the spike protein. They want to go beyond the existing static structures to calculate how the spike proteins move on the surface of the virus and how they change their shape – with a precision down to the size of an atom.
These calculations will reveal the tiniest details of the protein structure. But they are extremely complex. “We need the massive computing power of the supercomputers of the Max Planck Society”, explains Gerhard Hummer, Director at the Max Planck Institute of Biophysics.
With their dynamic model of the spike protein, the researchers hope to identify binding domains to which antibodies can reliably bind. Hummer and his team also hope to discover binding sites for inhibitors. They plan to compare these with the binding properties of existing drugs with the help of computers and thus identify active ingredients that can block the spike protein. “Of course, repurposing drugs that are already on the market is much faster than finding new active ingredients and testing them in lengthy clinical trials”, says Hummer.







